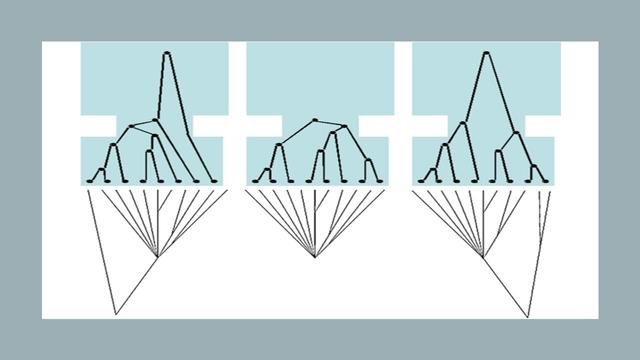
Excited to read this, I've been worried about the recurrence of these peaks in #PSMC graphs for years. Also, just on time before we submit one (a first for me). #PopulationGenetics #PopulationGenomics #MethodsMatter
https://bsky.app/profile/leonhilgers.bsky.social/post/3lhl5ti3nec25
(If Mastodon doesn't show the Bluesky post: "Avoidable false PSMC population size peaks occur across numerous studies" https://doi.org/10.1016/j.cub.2024.09.028)