Anyone there with experience on Labplot vs OriginLab and compatibility of the two (esp. Labplot’s ability to read OriginLab files) ??
#originlab #labplot #kde_labplot #labplot2 #physics #optics #photonics
#labplot
Some benefits of #LabPlot:
an interactive environment with a graphical user interface,
ease of use for non-programmers,
built-in tools for data analysis, statistics, regression, and curve fitting,
platform integration that may offer a better experience than running Python scripts,
possibility to run #Python, #Maxima, #R, #Octave and other languages inside LabPlot's computational notebooks.
Take a look at LabPlot's features:
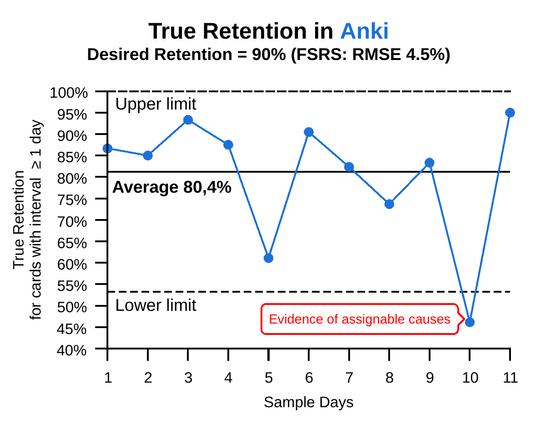
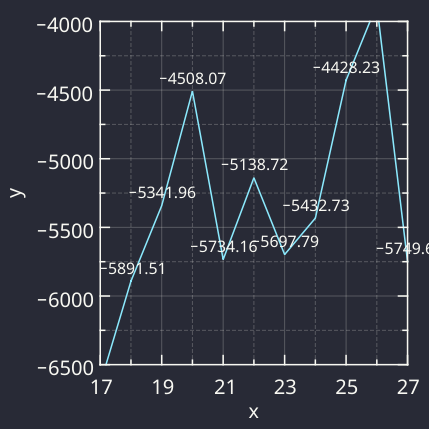
An example analysis of a #learning process with #LabPlot dev (P-chart).
@labplot@lemmy.kde.social @opensource
The process is off the target and has large variation. Variation always creates costs.
The point outside the limits is evidence that assignable causes with dominant effects are present and the process will behave UNPREDICTABLY.
'With an unpredictable process, PREDICTION IS FUTILE, but action may be taken to move the process closer to its full potential.' D.J. #Wheeler
Today I posted my first "Issue" / Bug Report to the amazing #FreeSoftware data analysis tool, #LabPlot . I am using it on Windows at work, to make nice graphs for my classes, and try to push it as hard as possible, to discover issues, and be able to make suggestions to improve usability.
So, I hope the issue I found will help the team improve the product, and I hope I will be able to give useful feature suggestions based on my experience.
@LabPlot at the service of #science and #researchers!
@labplot@lemmy.kde.social @opensource @openscience @organic_chemistry @ChemistryViews
#LabPlot is a FREE, open source and cross-platform Data Visualization and Analysis software.
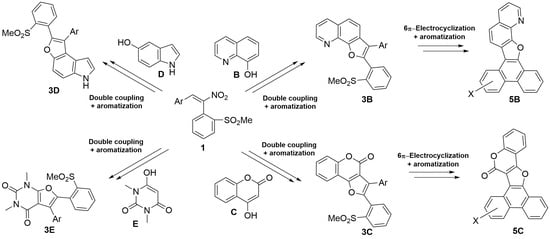
We're pleased to know that #LabPlot was used to perform calculations in this recent study on robust access to furo-fused #heteropolycycles:
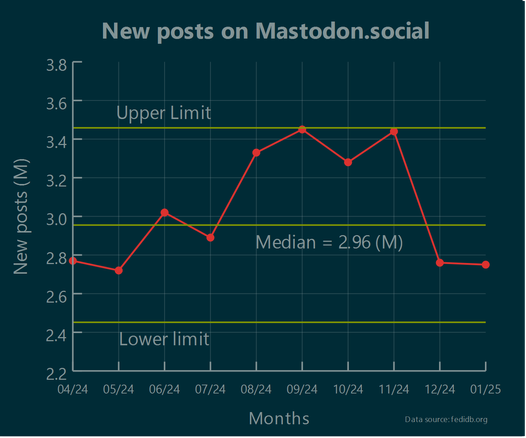
Is the number of new posts published on @Mastodon  growing or is it stable in the last few months?
growing or is it stable in the last few months?
How to visualize #server #metrics in #RealTime via #TCP in @LabPlot ?
@labplot@lemmy.kde.social @sysadmin@lemmy.world @sysadmin@lemmy.ml @cybersecurity
The purpose of this simple tutorial is not to position #LabPlot against dedicated applications, but rather to show how its "Live Data" functionality can be used to read and visualize data in real time.
https://docs.labplot.org/en/tutorials/live_data/tutorials_live_data_server_monitoring_via_tcp.html
@europesays @UnitedStates @labplot@lemmy.kde.social @dataisbeautiful
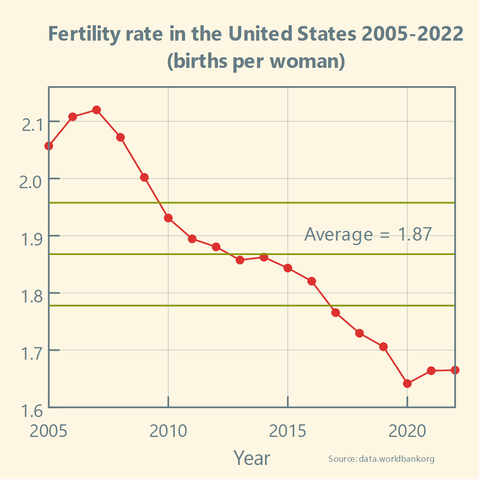
Has the #FertilityRate in the #US been stable over the past two decades? And how does it compare to the #EU?
Boosts appreciated! 
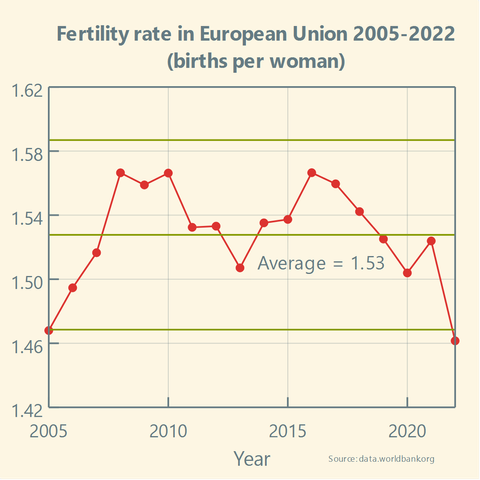
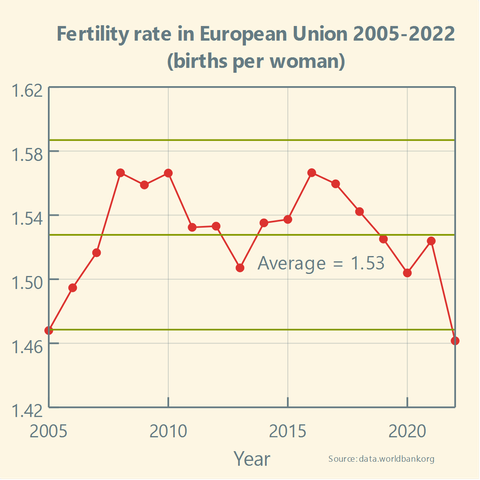
Has the #FertilityRate in the #EuropeanUnion been stable over the past two decades?
A simple #XmR chart available in @LabPlot [2.12dev] can be used to track the stability of any process.
Boosts appreciated! 
https://en.wikipedia.org/wiki/Shewhart_individuals_control_chart
It depends on the expected functionality. You can check the existing features here: https://labplot.org/features
We are currently working on expanding #LabPlot's functionality in these areas:
Live Data Analysis
#Python Scripting
Statistical Analysis
Quality Improvement Charts
#LabPlot is a FREE, open source and cross-platform Data Visualization and Analysis software accessible to everyone. In LabPlot your #data is yours alone!
In this short video you can learn how to quickly import your data into #LabPlot and visualize it.
Boosts appreciated! 
@opensource @openscience @alternativeto @9to5linux @omgubuntu
Seven simple questions for #DecisionMakers:
How big? How much? How many?
Compared to what?
Why not a rate?
Per what? The diabolical denominator.
How were things defined, counted or measured?
What was taken into account (what was controlled for)?
What else should have been taken into account (controlled for)?
#LabPlot Team Loves #FreeSoftwareDay!
@fsfe
@opensource @libre_software @libre_culture
Boosts appreciated! 
#LabPlot at the service of #science and #researchers!
Boosts appreciated! 
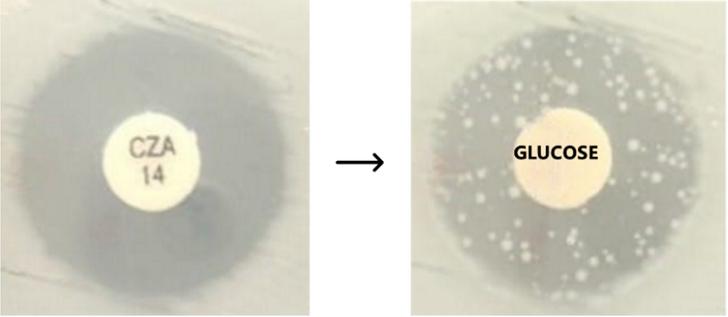
We're pleased to know that #LabPlot was used in this recent study on ceftazidime-avibactam (CZA) tolerance/persistence:
https://link.springer.com/article/10.1007/s10096-024-05005-4
#LabPlot at the service of #science and #researchers!
Boosts appreciated! 
It's rewarding to us to know that LabPlot was used for #DataVisualization in this recent study on oxidative and excitatory neurotoxic stresses:
https://www.imrpress.com/journal/FBL/30/1/10.31083/FBL25706/htm
Do you know why did the minus sign break up with the hyphen? Because it just couldn't handle the relationship—it was always too negative!
Boosts appreciated! 
Or, maybe, it's because we simply realized which minus sign symbol should be used in #LabPlot.
In the [dev] version we now use the minus sign symbol instead of the hyphen.
https://invent.kde.org/education/labplot/-/merge_requests/667
Today is the Data Privacy (Protection) Day! So let us remind you that in #LabPlot, an open-source data analysis and visualization software, Your Data is Yours!
@labplot@lemmy.kde.social @opensource @libre_software @privacy
Boosts appreciated!

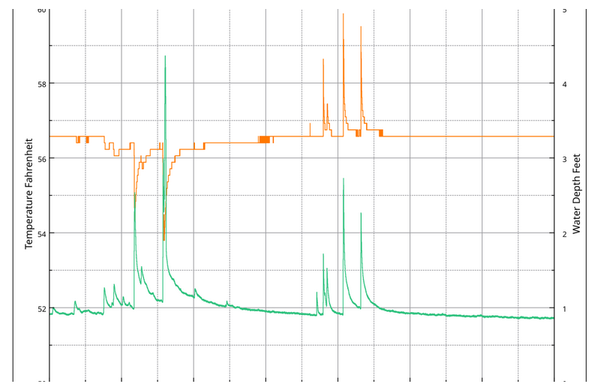
Would you like to know how #LabPlot, an open-source data analysis and visualization software, makes it easier to uncover the mysteries of #caves?
Then read this interesting article by Bill Gee:
https://www.mocavesandkarst.org/2023-stream-flow-project-report/
All images courtesy of Bill.
Boosts appreciated!

#LabPlot at the service of researchers.
It's rewarding to us to know that LabPlot was used for #DataVisualization in this recent study on metastatic breast cancer:
https://www.sciencedirect.com/science/article/pii/S0960977625000049
wegen Recherche zu #PeerTube_potentials
hab ich grad noch ein video #labplot gesehen ,
wollte ich schon länger mal ausprobieren .. zum Vergleich mit python-> matplotlib ..
https://tube.tchncs.de/w/i7djYpB38L1uRL5Fez2JCy
Abo-Kanal bei
https://tube.tchncs.de/c/s3nnet/videos
@s3nnet